This article was first published on Medium.
Have you worked with constrained resources (RAM and CPU) to accomplish large file/data parsing, especially a primitive file type like XML? This article discusses one such project where XML files of varying sizes (sometimes >10GB) were to be read, parsed and information from them to be stored in a Postgres database. This process was run every day as new files were uploaded to an online resource.
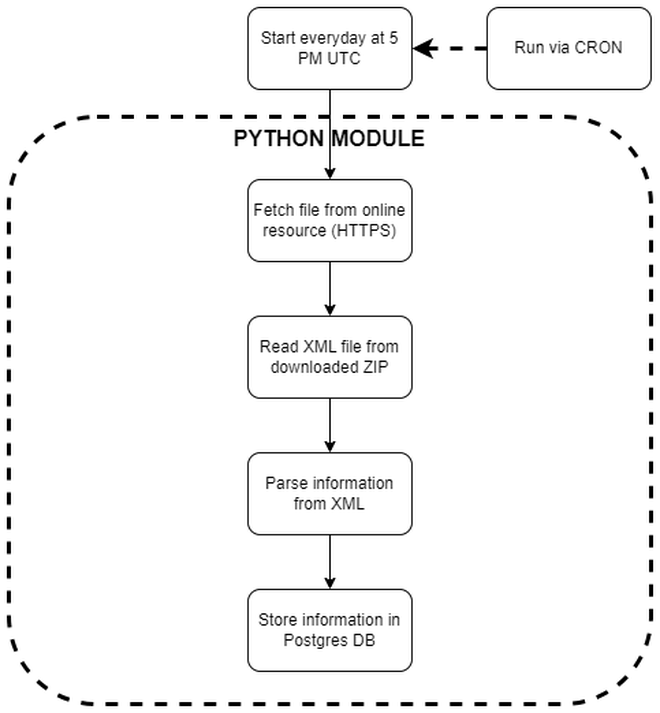
XML file layout #
The XML file layout is shown below. The number of n3 nodes could range from just a few to millions depending on the day when the file is downloaded. Many more sub-nodes exist within the n3 nodes and information from all is relevant; the sample shown is abridged for brevity. Nested nodes, and the context to which they belong are also important, i.e., the name node in n321 and n331 represent different pieces of information. Their parent node determines where in the database this information is placed.
<root dtd-version="1.0" date-produced="20131101">
<n1>DA</n1>
<n2>
<date>20250221</date>
</n2>
<n3s>
<n3>
<n31>
<n311>64347</n311>
<n312>1</n312>
...
<!-- more nodes here -->
</n31>
<n32>
<n321>
<name>Some Name</name>
...
<!-- more nodes here -->
</n321>
</n32>
<n33>
<n331>
<name>Some name</name>
...
<!-- more nodes here -->
</n331>
</n33>
<n34>
<n341>
<n3411>
<country>US</country>
...
<!-- more nodes here -->
</n3411>
<n3412>
<country>US</country>
...
<!-- more nodes here -->
</n3412>
<n342 lang="en">Some title</n342>
</n341>
...
<!-- more nodes here as n341 with different information -->
</n34>
</n3>
<n3>
<!-- data for the next n3 -->
</n3>
...
<!-- more nodes like n3 -->
</n3s>
Virtual machine (VM) #
The database is hosted on a virtual machine provisioned via Linode. With 16 GB RAM and 6 virtual CPUs (vCPUs), it has enough computing power to do a plethora of tasks. However, my naivete with the XML format, and its parsing via Python, rendered this virtual machine inadequate to perform the simple task outlined in the flowchart above. Specifically, earlier versions of the module would consume the entirety of RAM (and Swap memory) before the OS had to kill the process.
Implementation #
Python and the venerable lxml library were used to parse XML files, and asyncpg to interface with the Postgres database. The first version of the code was inefficient. It worked well on small files, where the file contents would fit into RAM. While developing, I tested this version with multiple files from the online source. To my (later) dismay, all of these files fit into RAM and the code worked, giving me false assurance that all files would be of similar sizes (+/- a few MBs), and everything would work just fine. It was not until a few months later, after checking the logs, that this assumption was challenged; a lot of Killed in the logs.
import subprocess
import asyncpg
import os
from pathlib import Path
from typing import Any
from lxml import etree
from src.models import N3 # Dataclass
async def connect_db() -> asyncpg.Connection:
connection = await asyncpg.connect(
user=POSTGRES_DB_USER,
password=POSTGRES_DB_PASS,
database=POSTGRES_DB_NAME,
host=POSTGRES_DB_HOST,
)
return connection
async def add_to_db(db_values: list[Any]):
connection = await connect_db()
await connection.copy_records_to_table(
table_name="table_name", records=db_values, columns=COLUMNS
)
await connection.close()
def read_xml_file(filename: str | Path) -> list[N3]:
n_root = etree.parse(filename)
n_3s = n_root.getroot().find("n3s")
list_n3: list[N3] = []
for n3 in n_3s.findall("n3"):
n_n31_rec = n3.find("n31")
n31_rec = create_n31_record(n_n31_rec)
n_n32_rec = n3.find("n32")
n32_rec = create_n32_record(n_n32_rec)
# continue with n33 and n32
list_n3.append(
N3(
n31=n31_rec,
n32=n32_rec,
n33=n33_rec,
n34=n34_rec,
)
)
return list_n3
async def process_one_xml_file(filename: str | Path) -> list[Any]:
xml_contents = read_xml_file(filename)
db_values = []
for xml_content in xml_contents:
# logic to get relevant information
db_value = get_relevant_info(xml_content)
db_values.append(tuple(db_value))
return db_values
async def process_one_link(link: str):
filename = os.path.basename(link)
# Wget the link
wget = subprocess.run(["wget", "--no-verbose", "--inet4-only", link])
# Unzip
unzip = subprocess.run(["unzip", filename, "-d", "xml_doc"])
# List XML files
files_in_folder = os.listdir("xml_doc")
for file in files_in_folder:
if ".xml" in file.lower():
db_values = await process_one_xml_file(os.path.join("xml_doc", file))
if db_values is None:
continue
await add_to_db(db_values)
# Remove everything
subprocess.run(["rm", filename])
subprocess.run(["rm", "-Rf", "xml_doc"])
Using this version, for a large file (~1.3GB in size, ~450,000 n3 nodes), successive HTOP views are shown in the picture below. As the file was loaded, RAM usage started increasing up to a point that it started creeping into Swap memory.
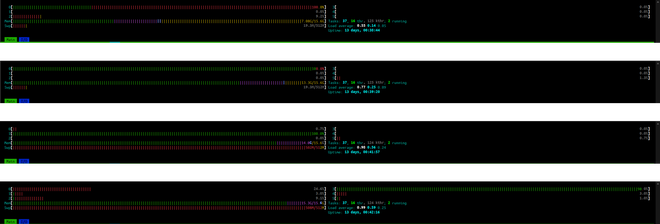
Two improvements were made to make the code efficient.
Using Python’s zipfile to remove redundant “unzip”ing at the OS level
#
In the first version, using the subprocess.run command, the zip file was being unzipped to read the XML file contents in function process_one_link. This was not the RAM killer, however, it was unnecessary. Python’s standard zipfile module is capable of reading the contents of individual files from the archive, without actually unzipping. The function was transformed as shown below. The .namelist() method provides a list of filenames within the archive. These can be filtered using list comprehension to get the .xml files.
import zipfile
async def process_one_link(link: str):
filename = os.path.basename(link)
# Wget the link
wget = subprocess.run(["wget", "--no-verbose", "--inet4-only", link])
with zipfile.ZipFile(filename) as pat_zip:
files = pat_zip.namelist()
xml_files = [x for x in files if ".xml" in x.lower()]
db_values = await process_one_xml_file(xml_files, pat_zip)
if db_values is None:
return
await add_to_db(db_values)
# ... rest of the function
In this implementation, the function signature of process_one_xml_file changes as well. What previously was taking just a str | Path as an input, now takes two arguments — a list of XML files within the archive and the zip archive itself. The function implementation changed slightly to incorporate using zipfile to open its contents without unzipping. zipfile.open reads bytes from the XML file within the archive; the output type of this operation is IO[bytes].
async def process_one_xml_file(
xml_files: list[str], zip_file: zipfile.ZipFile
) -> list[Any]:
db_values = []
for xml_file in xml_files:
with zip_file.open(xml_file) as file:
xml_contents = read_xml_file(file)
root_logger.info("Successfully read XML file")
# ... rest of the implementation
return db_values
Using iterparse instead of loading entire XML in RAM using parse #
Previously, in read_xml_file, the contents of the XML file were loaded in memory using etree.parse. It then parsed the n3s node to iterate over all n3 nodes within it using the findall method. An alternative to parse is to use event-based parsing of XML files using iterparse. Event-based parsing is where “start” and “end” events of a XML tag are recorded, and only the data between that tag are loaded into memory. A remarkable advantage of event-based parsing is that the events can be cleared, i.e., data from one tag can be removed to free up memory. With this, the read_xml_file function changed. The findall for loop was changed to an iterparse for loop. A single call of .clear() cleared the current n3 node’s data from memory because its contents were already parsed and stored in list_n3.
def read_xml_file(file_contents: IO[bytes]) -> list[N3]:
list_n3: list[N3] = []
for _, n3 in etree.iterparse(
file_contents, tag="n3", encoding="utf-8", recover=True
):
# rest of the for loop stays the same
n3.clear()
return list_n3
With these two changes, running out of RAM has never been a problem. When the same file (~1.3 GB and ~450,000 n3 nodes) was processed with the new logic, i.e., with iterparse and .clear(), RAM increases steadily as new data is being added into list_n3.
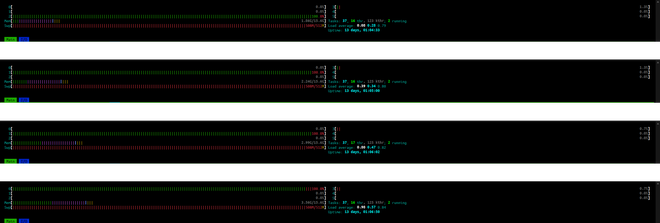
Conclusion #
For an XML veteran and a heavy lxml user, the findings from this article may be obvious. However, it took failures in the pipeline for me to know and understand the problem. I was tempted to “throw more hardware” to get to a solution, which would have slowly started chipping away at our bottom line. Even with inexpensive cloud providers like Linode provisioning VMs with additional RAM can add up costs quickly. It is always better to find a different way to do things, rather than adding more hardware, may it be using a different library or using a different programming language altogether.
